ATAC-seq has been a relatively sequencing technology in recent years. What exactly is the ATAC-seq technology? The full name of ATAC-seq is Assay for Transposase Accessible Chromatin using sequencing, a technique for studying the chromatin accessible by transposase using sequencing. This technique cleaves a nuclear chromatin region that is open at a specific time and space by a transposase, thereby obtaining regulatory sequences of all active transcriptions in the genome at that specific time and space. This technology can obtain real-time genome-wide activity regulatory sequence information with only a small number of cells, and is widely used in transcription factor binding analysis, nucleosome localization, and distribution of active regulatory elements, and has broad application prospects in the field of epigenetic mechanism research. .
So what is the popularity of the ATAC-seq study? Let's take a look at the articles related to the ATAC-seq technology. According to the latest statistics, the number of articles published using ATAC-seq technology has grown exponentially; among them, 302 and its magazines, 215 articles of Nature and its magazines, and 82 articles of Science and its magazines. It is conceivable that there is much fire in the ATAC-seq technology. In this large research background, what kind of ATAC-seq service does Yunxu Biosystem provide as a professional sequencing service company?
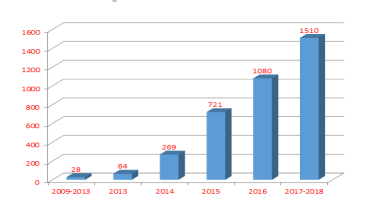
Number of articles published by ATAC-seq
Cloud Sequence Creature - ATAC-seq One ​​Stop Shop
One-stop service process

2. Technical advantages
Fast: the experimental process is streamlined and time consuming
Micro: small amount of cells, suitable for clinical samples
Accurate: good repeatability, high consistency with similar technologies and similar sequencing platforms
Comprehensive: access to a large amount of information (full genome regulation activity map, full transcription factor binding map)
3. ATAC-seq information analysis
Standard analysis: Peak detection, Peak length detection, Peak depth distribution, Peak-related genes, differential peak detection between samples, differential peak-related genes between samples, GO and KEGG enrichment analysis of differential peak-related genes between samples
Advanced information analysis: nucleosome localization analysis, transcription factor binding analysis (requires customers to provide TF names of interest), genome-wide activity map
Joint analysis: association analysis between ATAC-Seq and RNA-Seq, correlation analysis between ATAC-Seq and ChIP-Seq
Cloud sequence biology - ATAC-seq related article ideas summary
1. Association analysis between ATAC-Seq and RNA-Seq
The ATAC-seq results, we studied the genes that are transcribed under this spatiotemporal condition and some sequences of homeopathic regulatory elements, then we can analyze these genes. Combining the results of RNA-seq, looking at some of the abundance of DNA sequences detected on ATAC, whether the corresponding transcript expression is also increased, can also find the corresponding upstream regulatory sequences of transcript-related genes, so that the overall analysis from DNA The process of RNA transcription, and further analysis of gene function, combined with experimental phenotypes, to clarify the process of apparent regulation-expression-function-phenotype.
Case analysis:
EGRINs (EnvironmentalGene Regulatory Influence Networks) in Rice That Function in the Response to Water Deficit, High Temperature, and Agricultural Environments OPEN. The Plant Cell, IF: 8.228
This article is a classic case of joint analysis using ATAC-seq and RNA-seq. The authors selected five different rice varieties and heat treated and dried them separately to simulate the two stress environments of high temperature and water shortage. Furthermore, RNA-seq detection of RNA expression differences and ATAC-seq were used to detect differences in chromatin open regions in five rice cultivars treated with this stress. Finally, the ATAC-seq data and RNA-seq data are combined with the transcription information of the transcription factor. Construction of a regulatory network map of plant-stressed transcription factor-target genes and the search for key transcription factor-target gene modules that predict key transcription factor activity. Complete studies of transcription factors involved in plant response in response to conditions.
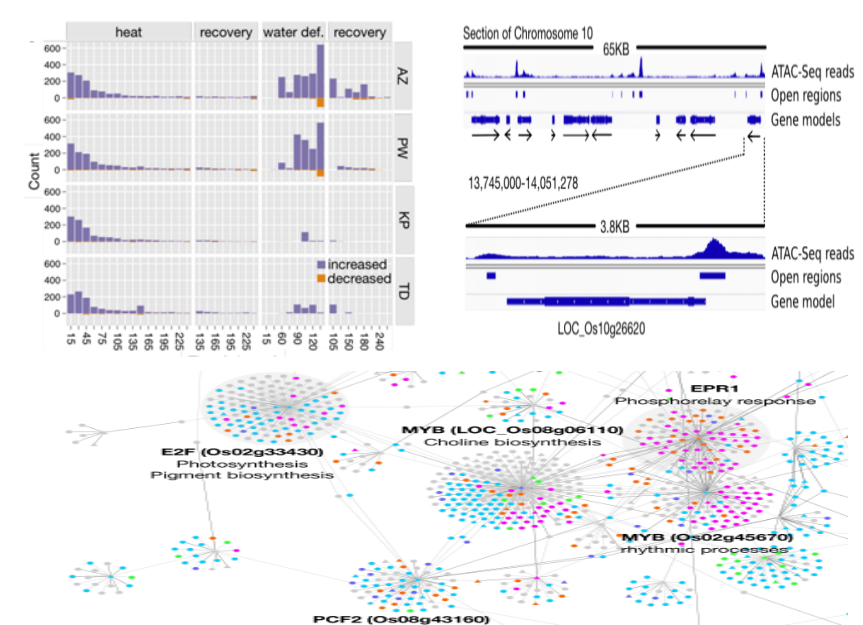
Joint analysis of ATAC-seq and RNA-seq
2. Joint analysis of ATAC-seq and ChIP-seq
In the ChIP experiment, many times we used to study those genes regulated by transcription factors. So, do you not need to do ChIP experiments after ATAC sequencing? In fact, it is not like transcriptome sequencing, you can know the up-and-down of gene expression, but we still need to use qPCR for molecular biology verification. After ATAC sequencing, ChIP-seq is also needed for further verification. The sequencing results of ChIP are used to further verify some of the transcription factor binding regions predicted by ATAC.
Case analysis:
Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility.Cell, IF:31.398
This article examines the background mechanism of human small cell lung cancer to promote cancer spread and metastasis by comparing the differences between primary and metastatic small cell lung cancer cells. First, using ATAC-seq, it was found that NFI family transcription factors are enriched in the open chromatin open sites, indicating that NFI family transcription factors play an important role in regulating tumor cell metastasis. Further ChIP-seq, combined analysis of the two sequencing results, found that the number of copies of Nfib was increased in the high open site of chromatin, and Nfib was highly expressed in invasive primary tumors and metastatic tumors, Nfib expression The function of maintaining chromatin and opening of the distal regulatory region and promoting the expression of neural genes indicates that Nfib plays an important role in promoting cancer cell proliferation and migration.

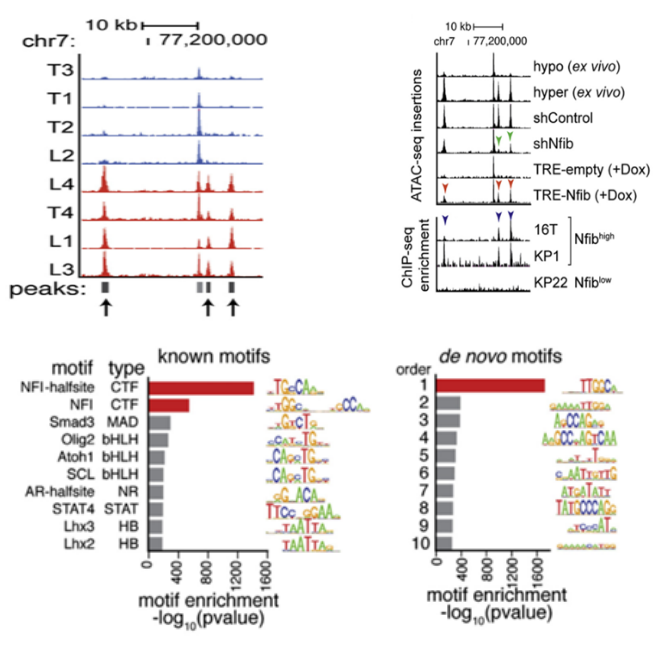
Joint analysis of ATAC-seq and ChIP-seq
Cloud order biology related recommendations.
ChIP sequencing
Whole transcriptome sequencing
Circular RNA sequencing
lncRNA sequencing
mRNA sequencing
miRNA sequencing
Wonderful review in the past
Daniel tells you how to study lncRNA methylation
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - non-coding RNA articles
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - virus articles
The latest cloud sequence biology m6A "RNA methylation" research summary - cancer articles
The first domestic 10+ M6A RNA methylation sequencing article---Cloud Order Bio-Help!
Yunxu Bio exclusive M5C RNA methylation sequencing
2018 National Nature Research Hotspot 2: In-depth analysis of RNA methylation research 20 points of RNA methylation in the article, heat and strength coexist
Genome research: There are huge differences in methylation modification of different RNA m5C
Plant Cell: Arabidopsis found a new m6A RNA demethylation modifying enzyme
Molecular cell: a new method for m1A RNA methylation sequencing--m1A-MAP
Hepatology: m6A RNA methylase METTL3 promotes the development of liver cancer
Nature Heavy: m6A RNA methylation is involved in the key aspects of hematopoietic stem cell development
Cancer cell: Professor He Chuan discovered the important function of m6A RNA methylation
Genome Biology: Using RNA sequencing to publish RNA methylation 10 points
RNA methylation book (2) m6A methylation and MeRIP sequencing
m5C RNA methylation research method
Shanghai Yunxu Biological Technology Co., Ltd.
Shanghai Cloud-seq Biotech Co., Ltd.
Address: 3rd Floor, Building 20, No. 518, Zhangzhu Road, Songjiang District, Shanghai 
Telephone Fax Website:
mailbox:
Calcium Acetate Anhydrous,Calcium Acetate Anhydrous Powder,Anhydrous Acetate Calcium,Acetic Acid Calcium Salt
Wuxi Yangshan Biochemical Co.,Ltd. , https://www.salesacetates.com